Planetary Computer¶
Do you know Planetary Computer?
Here is a new level!
Level 8: spyndex + Planetary Computer!
Remember to install spyndex and planetary-computer!
[ ]:
!pip install -U spyndex planetary-computer stackstac
And remember to configure Planetary Computer if you have an API key! (If not, that’s okay, just omit this step!)
[ ]:
!planetarycomputer configure
Now, let’s start!
We are going to partially recreate this Sentinel-2 Planetary Computer Tutorial. But now we are going to use spyndex to compute some Spectral Indices!
First, import everything we need:
[1]:
import spyndex
import stackstac
import planetary_computer as pc
import numpy as np
import xarray as xr
import matplotlib.pyplot as plt
from pystac_client import Client
Let’s open the catalog!
[2]:
catalog = Client.open("https://planetarycomputer.microsoft.com/api/stac/v1")
Define the IDs of the images we are goingto use:
[3]:
ids = [
"S2A_MSIL2A_20200616T141741_R010_T20LQR_20200822T232052",
"S2B_MSIL2A_20190617T142049_R010_T20LQR_20201006T032921",
"S2B_MSIL2A_20180712T142039_R010_T20LQR_20201011T150557",
"S2B_MSIL2A_20170727T142039_R010_T20LQR_20210210T153028",
"S2A_MSIL2A_20160627T142042_R010_T20LQR_20210211T234456",
]
Search for those IDs in the Sentinel-2 L2A Collection:
[4]:
search = catalog.search(collections = ["sentinel-2-l2a"], ids = ids)
And sign them with the Planetary Computer API!
[5]:
items = [pc.sign(item).to_dict() for item in search.get_items()]
Now, let’s use stackstac to create a dask.DataArray from the images!
[6]:
S2 = stackstac.stack(
items,
epsg = 32619,
resolution = 500,
assets = ["B04", "B05", "B06", "B07", "B08"],
chunksize = 256,
).where(lambda x: x > 0, other = np.nan)
Let’s round the time to a daily time scale:
[7]:
S2 = S2.assign_coords(time = lambda x: x.time.dt.round("D"))
Remember also to scale the collection!
[8]:
S2 = S2 / 10000
And now let’s check what we have here…
[9]:
S2
[9]:
<xarray.DataArray 'stackstac-e6141dd496554095669cb9f3981a0a12' (time: 5, band: 5, y: 227, x: 226)>
dask.array<truediv, shape=(5, 5, 227, 226), dtype=float64, chunksize=(1, 1, 227, 226), chunktype=numpy.ndarray>
Coordinates: (12/44)
* time (time) datetime64[ns] 2016-06-28...
id (time) <U54 'S2A_MSIL2A_20160627...
* band (band) <U3 'B04' 'B05' ... 'B08'
* x (x) float64 1.362e+06 ... 1.474e+06
* y (y) float64 -9.075e+05 ... -1.02...
s2:snow_ice_percentage float64 0.0
... ...
gsd (band) int64 10 20 20 20 10
proj:bbox object {699960.0, 809760.0, 8990...
common_name (band) <U7 'red' ... 'nir'
center_wavelength (band) float64 0.665 ... 0.842
full_width_half_max (band) float64 0.038 ... 0.145
epsg int64 32619- time: 5
- band: 5
- y: 227
- x: 226
- dask.array<chunksize=(1, 1, 227, 226), meta=np.ndarray>
Array Chunk Bytes 9.79 MiB 400.80 kiB Shape (5, 5, 227, 226) (1, 1, 227, 226) Count 151 Tasks 25 Chunks Type float64 numpy.ndarray 5 1 226 227 5 - time(time)datetime64[ns]2016-06-28 ... 2020-06-17
array(['2016-06-28T00:00:00.000000000', '2017-07-28T00:00:00.000000000', '2018-07-13T00:00:00.000000000', '2019-06-18T00:00:00.000000000', '2020-06-17T00:00:00.000000000'], dtype='datetime64[ns]') - id(time)<U54'S2A_MSIL2A_20160627T142042_R010...
array(['S2A_MSIL2A_20160627T142042_R010_T20LQR_20210211T234456', 'S2B_MSIL2A_20170727T142039_R010_T20LQR_20210210T153028', 'S2B_MSIL2A_20180712T142039_R010_T20LQR_20201011T150557', 'S2B_MSIL2A_20190617T142049_R010_T20LQR_20201006T032921', 'S2A_MSIL2A_20200616T141741_R010_T20LQR_20200822T232052'], dtype='<U54') - band(band)<U3'B04' 'B05' 'B06' 'B07' 'B08'
array(['B04', 'B05', 'B06', 'B07', 'B08'], dtype='<U3')
- x(x)float641.362e+06 1.362e+06 ... 1.474e+06
array([1361500., 1362000., 1362500., ..., 1473000., 1473500., 1474000.])
- y(y)float64-9.075e+05 -9.08e+05 ... -1.02e+06
array([ -907500., -908000., -908500., ..., -1019500., -1020000., -1020500.])
- s2:snow_ice_percentage()float640.0
array(0.)
- s2:dark_features_percentage(time)float640.06314 0.1075 ... 0.06388 0.06595
array([0.063142, 0.107498, 0.063019, 0.063882, 0.065949])
- s2:mean_solar_zenith(time)float6440.32 37.77 39.68 40.04 40.03
array([40.31880089, 37.76587633, 39.68319946, 40.04278651, 40.03236022])
- s2:thin_cirrus_percentage(time)float640.000793 0.001042 ... 0.00078
array([0.000793, 0.001042, 0.001005, 0.000916, 0.00078 ])
- s2:mean_solar_azimuth(time)float6437.17 42.63 39.26 36.43 36.43
array([37.17374055, 42.63485595, 39.26341594, 36.43317178, 36.42928038])
- s2:granule_id(time)<U62'S2A_OPER_MSI_L2A_TL_ESRI_202102...
array(['S2A_OPER_MSI_L2A_TL_ESRI_20210211T234459_A005298_T20LQR_N02.12', 'S2B_OPER_MSI_L2A_TL_ESRI_20210210T153031_A002038_T20LQR_N02.12', 'S2B_OPER_MSI_L2A_TL_ESRI_20201011T150558_A007043_T20LQR_N02.12', 'S2B_OPER_MSI_L2A_TL_ESRI_20201006T032923_A011905_T20LQR_N02.12', 'S2A_OPER_MSI_L2A_TL_ESRI_20200822T232055_A026033_T20LQR_N02.12'], dtype='<U62') - s2:datatake_id(time)<U34'GS2A_20160627T142042_005298_N02...
array(['GS2A_20160627T142042_005298_N02.12', 'GS2B_20170727T142039_002038_N02.12', 'GS2B_20180712T142039_007043_N02.12', 'GS2B_20190617T142049_011905_N02.12', 'GS2A_20200616T141741_026033_N02.12'], dtype='<U34') - s2:high_proba_clouds_percentage(time)float640.000554 0.00069 ... 0.000156 7e-05
array([5.54e-04, 6.90e-04, 3.88e-04, 1.56e-04, 7.00e-05])
- s2:generation_time(time)<U24'2021-02-11T23:44:56.943Z' ... '...
array(['2021-02-11T23:44:56.943Z', '2021-02-10T15:30:28.983Z', '2020-10-11T15:05:57.242Z', '2020-10-06T03:29:21.378Z', '2020-08-22T23:20:52.991Z'], dtype='<U24') - s2:nodata_pixel_percentage(time)float640.0 0.0 0.0 1.3e-05 3e-06
array([0.0e+00, 0.0e+00, 0.0e+00, 1.3e-05, 3.0e-06])
- proj:epsg()int6432720
array(32720)
- s2:degraded_msi_data_percentage()float640.0
array(0.)
- s2:datatake_type()<U8'INS-NOBS'
array('INS-NOBS', dtype='<U8') - s2:saturated_defective_pixel_percentage()float640.0
array(0.)
- sat:relative_orbit()int6410
array(10)
- s2:datastrip_id(time)<U64'S2A_OPER_MSI_L2A_DS_ESRI_202102...
array(['S2A_OPER_MSI_L2A_DS_ESRI_20210211T234459_S20160627T142038_N02.12', 'S2B_OPER_MSI_L2A_DS_ESRI_20210210T153031_S20170727T142105_N02.12', 'S2B_OPER_MSI_L2A_DS_ESRI_20201011T150558_S20180712T142035_N02.12', 'S2B_OPER_MSI_L2A_DS_ESRI_20201006T032923_S20190617T142044_N02.12', 'S2A_OPER_MSI_L2A_DS_ESRI_20200822T232055_S20200616T142338_N02.12'], dtype='<U64') - constellation()<U10'Sentinel 2'
array('Sentinel 2', dtype='<U10') - platform(time)<U11'Sentinel-2A' ... 'Sentinel-2A'
array(['Sentinel-2A', 'Sentinel-2B', 'Sentinel-2B', 'Sentinel-2B', 'Sentinel-2A'], dtype='<U11') - instruments()<U3'msi'
array('msi', dtype='<U3') - eo:cloud_cover(time)float640.00218 0.002621 ... 0.001108
array([0.00218 , 0.002621, 0.001831, 0.001523, 0.001108])
- s2:water_percentage(time)float640.4949 0.4862 0.497 0.5117 0.5117
array([0.494935, 0.48617 , 0.496953, 0.511657, 0.511657])
- sat:orbit_state()<U10'descending'
array('descending', dtype='<U10') - s2:medium_proba_clouds_percentage(time)float640.000833 0.000889 ... 0.000259
array([0.000833, 0.000889, 0.000438, 0.000451, 0.000259])
- s2:product_type()<U7'S2MSI2A'
array('S2MSI2A', dtype='<U7') - s2:unclassified_percentage(time)float640.007976 0.02441 ... 0.006699
array([0.007976, 0.024409, 0.005617, 0.005365, 0.006699])
- s2:processing_baseline()<U5'02.12'
array('02.12', dtype='<U5') - s2:vegetation_percentage(time)float6499.19 98.65 98.77 99.16 99.25
array([99.192262, 98.648018, 98.77103 , 99.161118, 99.248123])
- s2:mgrs_tile()<U5'20LQR'
array('20LQR', dtype='<U5') - s2:not_vegetated_percentage(time)float640.2395 0.7313 0.6615 0.2565 0.1665
array([0.239508, 0.731285, 0.661547, 0.256456, 0.166456])
- s2:reflectance_conversion_factor(time)float640.9679 0.9691 0.9674 0.9695 0.9695
array([0.96787441, 0.96909248, 0.96743467, 0.9694882 , 0.96954851])
- s2:cloud_shadow_percentage(time)float640.0 0.0 0.0 3e-06 0.0
array([0.e+00, 0.e+00, 0.e+00, 3.e-06, 0.e+00])
- s2:product_uri(time)<U65'S2A_MSIL2A_20160627T142042_N021...
array(['S2A_MSIL2A_20160627T142042_N0212_R010_T20LQR_20210211T234456.SAFE', 'S2B_MSIL2A_20170727T142039_N0212_R010_T20LQR_20210210T153028.SAFE', 'S2B_MSIL2A_20180712T142039_N0212_R010_T20LQR_20201011T150557.SAFE', 'S2B_MSIL2A_20190617T142049_N0212_R010_T20LQR_20201006T032921.SAFE', 'S2A_MSIL2A_20200616T141741_N0212_R010_T20LQR_20200822T232052.SAFE'], dtype='<U65') - title(band)<U36'Band 4 - Red - 10m' ... 'Band 8...
array(['Band 4 - Red - 10m', 'Band 5 - Vegetation red edge 1 - 20m', 'Band 6 - Vegetation red edge 2 - 20m', 'Band 7 - Vegetation red edge 3 - 20m', 'Band 8 - NIR - 10m'], dtype='<U36') - gsd(band)int6410 20 20 20 10
array([10, 20, 20, 20, 10])
- proj:bbox()object{699960.0, 809760.0, 8990200.0, ...
array({699960.0, 809760.0, 8990200.0, 9100000.0}, dtype=object) - common_name(band)<U7'red' 'rededge' ... 'rededge' 'nir'
array(['red', 'rededge', 'rededge', 'rededge', 'nir'], dtype='<U7')
- center_wavelength(band)float640.665 0.704 0.74 0.783 0.842
array([0.665, 0.704, 0.74 , 0.783, 0.842])
- full_width_half_max(band)float640.038 0.019 0.018 0.028 0.145
array([0.038, 0.019, 0.018, 0.028, 0.145])
- epsg()int6432619
array(32619)
That’s INCREDIBLE! We have our images as a dask.DataArray!
And… if you remember the Dask tutorial, computing Spectral Indices with spyndex and dask is super simple!
Let’s compute the NDVI and the IRECI:
[14]:
idx = spyndex.computeIndex(
index = ["NDVI","IRECI"],
params = {
"N": S2.sel(band = "B08"),
"R": S2.sel(band = "B04"),
"RE1": S2.sel(band = "B05"),
"RE2": S2.sel(band = "B06"),
"RE3": S2.sel(band = "B07"),
}
)
Now, let’s check our result:
[15]:
idx
[15]:
<xarray.DataArray 'stackstac-e6141dd496554095669cb9f3981a0a12' (index: 2, time: 5, y: 227, x: 226)> dask.array<concatenate, shape=(2, 5, 227, 226), dtype=float64, chunksize=(1, 1, 227, 226), chunktype=numpy.ndarray> Coordinates: * time (time) datetime64[ns] 2016-06-28 2017-07-28 ... 2020-06-17 * x (x) float64 1.362e+06 1.362e+06 1.362e+06 ... 1.474e+06 1.474e+06 * y (y) float64 -9.075e+05 -9.08e+05 -9.085e+05 ... -1.02e+06 -1.02e+06 * index (index) <U5 'NDVI' 'IRECI'
- index: 2
- time: 5
- y: 227
- x: 226
- dask.array<chunksize=(1, 1, 227, 226), meta=np.ndarray>
Array Chunk Bytes 3.91 MiB 400.80 kiB Shape (2, 5, 227, 226) (1, 1, 227, 226) Count 226 Tasks 10 Chunks Type float64 numpy.ndarray 2 1 226 227 5 - time(time)datetime64[ns]2016-06-28 ... 2020-06-17
array(['2016-06-28T00:00:00.000000000', '2017-07-28T00:00:00.000000000', '2018-07-13T00:00:00.000000000', '2019-06-18T00:00:00.000000000', '2020-06-17T00:00:00.000000000'], dtype='datetime64[ns]') - x(x)float641.362e+06 1.362e+06 ... 1.474e+06
array([1361500., 1362000., 1362500., ..., 1473000., 1473500., 1474000.])
- y(y)float64-9.075e+05 -9.08e+05 ... -1.02e+06
array([ -907500., -908000., -908500., ..., -1019500., -1020000., -1020500.])
- index(index)<U5'NDVI' 'IRECI'
array(['NDVI', 'IRECI'], dtype='<U5')
A dask.DataArray! You know what that means… We have to compute the indices!
[16]:
idx = idx.compute()
And finally, you can plot the indices! Here the NDVI:
[17]:
idx.sel(index = "NDVI").plot.imshow(col = "time",cmap = "viridis_r")
[17]:
<xarray.plot.facetgrid.FacetGrid at 0x7f5fc7c69a90>
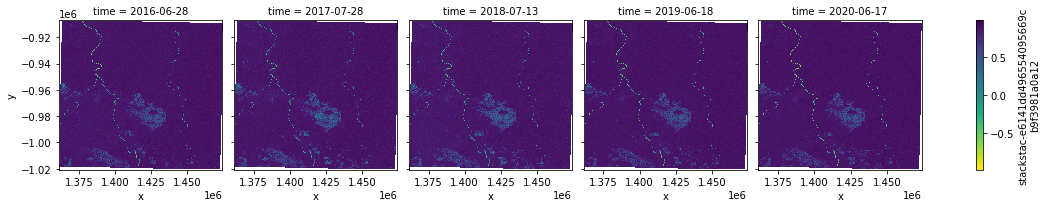
And here the IRECI:
[18]:
idx.sel(index = "IRECI").plot.imshow(col = "time",cmap = "viridis_r")
[18]:
<xarray.plot.facetgrid.FacetGrid at 0x7f5fc41ec730>
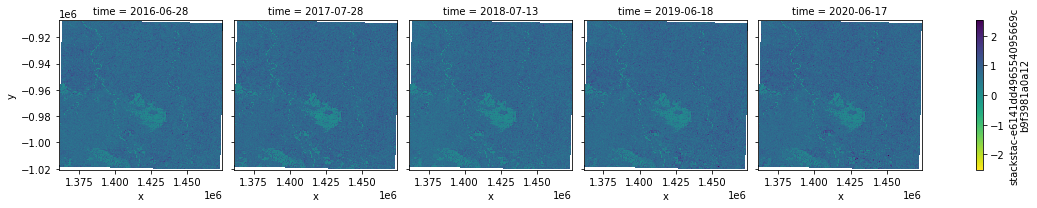
Wow! :D
