Pandas¶
After passing levels 1 and 2, you are ready to start this: Level 3 - spyndex + pandas!
Remember to install spyndex!
[ ]:
!pip install -U spyndex
Now, let’s start!
First, import spyndex and pandas:
[31]:
import spyndex
import pandas as pd
pandas.Series¶
We have all worked with pandas. Well, spyndex also works with pandas so you can continue using it! :)
Let’s use a pandas.DataFrame that is stored in the spyndex datasets: spectral:
[59]:
df = spyndex.datasets.open("spectral")
Each column of this dataset is the Surface Reflectance from Landsat 8 for 3 different classes. The samples were taken over Oporto:
[33]:
df.head()
[33]:
| SR_B1 | SR_B2 | SR_B3 | SR_B4 | SR_B5 | SR_B6 | SR_B7 | ST_B10 | class | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.089850 | 0.100795 | 0.132227 | 0.165764 | 0.269054 | 0.306206 | 0.251949 | 297.328396 | Urban |
| 1 | 0.073859 | 0.086990 | 0.124404 | 0.160979 | 0.281264 | 0.267596 | 0.217917 | 297.107934 | Urban |
| 2 | 0.072938 | 0.086028 | 0.120994 | 0.140203 | 0.284220 | 0.258384 | 0.200098 | 297.436064 | Urban |
| 3 | 0.087733 | 0.103916 | 0.135981 | 0.163976 | 0.254479 | 0.259580 | 0.216735 | 297.203638 | Urban |
| 4 | 0.090593 | 0.109306 | 0.150350 | 0.181260 | 0.269535 | 0.273234 | 0.219554 | 297.097680 | Urban |
Here you can see the classes stored in the class column:
[34]:
df["class"].unique()
[34]:
array(['Urban', 'Water', 'Vegetation'], dtype=object)
Each column of the data frame is a pandas.Series data type:
[35]:
type(df["SR_B2"])
[35]:
pandas.core.series.Series
Well, we can use that to compute Spectral Indices with spyndex!
Since we have vegetation, water and urban classes, let’s compute 3 different indices, each one highlighting an specific class: NDVI, NDWI and NDBI:
[36]:
spyndex.indices.NDVI
[36]:
NDVI: Normalized Difference Vegetation Index (attributes = ['bands', 'contributor', 'date_of_addition', 'formula', 'long_name', 'reference', 'short_name', 'type'])
[37]:
spyndex.indices.NDWI
[37]:
NDWI: Normalized Difference Water Index (attributes = ['bands', 'contributor', 'date_of_addition', 'formula', 'long_name', 'reference', 'short_name', 'type'])
[38]:
spyndex.indices.NDBI
[38]:
NDBI: Normalized Difference Built-Up Index (attributes = ['bands', 'contributor', 'date_of_addition', 'formula', 'long_name', 'reference', 'short_name', 'type'])
What bands do we need?
[39]:
spyndex.indices.NDVI.bands
[39]:
('N', 'R')
[40]:
spyndex.indices.NDWI.bands
[40]:
('G', 'N')
[41]:
spyndex.indices.NDBI.bands
[41]:
('S1', 'N')
Green, Red, NIR and SWIR1 bands… easy!
[42]:
parameters = {
"G": df["SR_B3"],
"R": df["SR_B4"],
"N": df["SR_B5"],
"S1": df["SR_B6"],
}
With our dict of parameters ready we can compute the indices!
[43]:
idx = spyndex.computeIndex(["NDVI","NDWI","NDBI"],parameters)
And, what’s the data type of the result?
[44]:
print(f"idx type: {type(idx)}")
idx type: <class 'pandas.core.frame.DataFrame'>
That’s right! A pandas.DataFrame! Why? Because each computed spectral index is now a column (pandas.Series) of a new dataframe:
[45]:
idx.head()
[45]:
| NDVI | NDWI | NDBI | |
|---|---|---|---|
| 0 | 0.237548 | -0.340973 | 0.064584 |
| 1 | 0.271989 | -0.386671 | -0.024902 |
| 2 | 0.339326 | -0.402815 | -0.047615 |
| 3 | 0.216278 | -0.303482 | 0.009923 |
| 4 | 0.195821 | -0.283852 | 0.006815 |
If you want them diectly on the original dataframe as new columns, you just have to play with the code a little bit!
[60]:
indicesToCompute = ["NDVI","NDWI","NDBI"]
df[indicesToCompute] = spyndex.computeIndex(indicesToCompute,parameters)
Now, if you check you original dataframe, you should have the new indices there!
[61]:
df.head()
[61]:
| SR_B1 | SR_B2 | SR_B3 | SR_B4 | SR_B5 | SR_B6 | SR_B7 | ST_B10 | class | NDVI | NDWI | NDBI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.089850 | 0.100795 | 0.132227 | 0.165764 | 0.269054 | 0.306206 | 0.251949 | 297.328396 | Urban | 0.237548 | -0.340973 | 0.064584 |
| 1 | 0.073859 | 0.086990 | 0.124404 | 0.160979 | 0.281264 | 0.267596 | 0.217917 | 297.107934 | Urban | 0.271989 | -0.386671 | -0.024902 |
| 2 | 0.072938 | 0.086028 | 0.120994 | 0.140203 | 0.284220 | 0.258384 | 0.200098 | 297.436064 | Urban | 0.339326 | -0.402815 | -0.047615 |
| 3 | 0.087733 | 0.103916 | 0.135981 | 0.163976 | 0.254479 | 0.259580 | 0.216735 | 297.203638 | Urban | 0.216278 | -0.303482 | 0.009923 |
| 4 | 0.090593 | 0.109306 | 0.150350 | 0.181260 | 0.269535 | 0.273234 | 0.219554 | 297.097680 | Urban | 0.195821 | -0.283852 | 0.006815 |
Beautiful! Right?
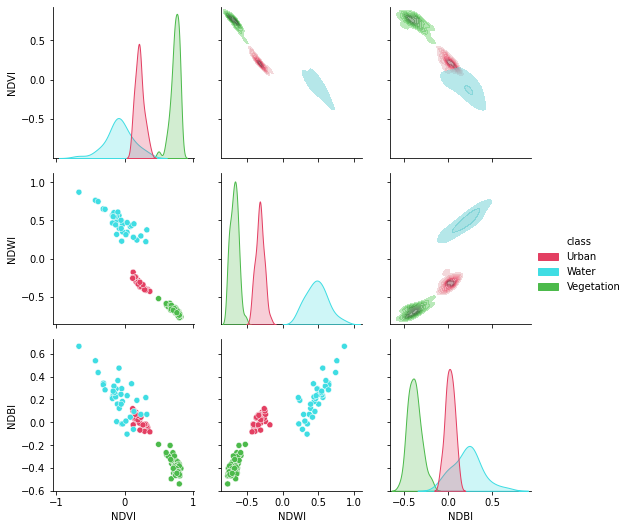
Now, just for the sake of life, let’s make some visualizations!
[50]:
import seaborn as sns
import matplotlib.pyplot as plt
Define some colors for each one of the classes:
[49]:
colors = ["#E33F62","#3FDDE3","#4CBA4B"]
Now, let’s create a gorgeous pair grid!
[64]:
plt.figure(figsize = (15,15))
g = sns.PairGrid(df[['NDVI', 'NDWI', 'NDBI','class']],hue = "class",palette = sns.color_palette(colors))
g.map_lower(sns.scatterplot)
g.map_upper(sns.kdeplot,fill = True,alpha = .5)
g.map_diag(sns.kdeplot,fill = True)
g.add_legend()
plt.show()
<Figure size 1080x1080 with 0 Axes>